Aplysia Transcriptome Assembly
This website for this Aplysia transcriptome was developed by the Institute for Genome Sciences at the University of Maryland, Baltimore.
This assembly was produced based on Illumina RNAseq of Aplysia mRNA from CNS and peripheral tissues using the Trinity assembler.
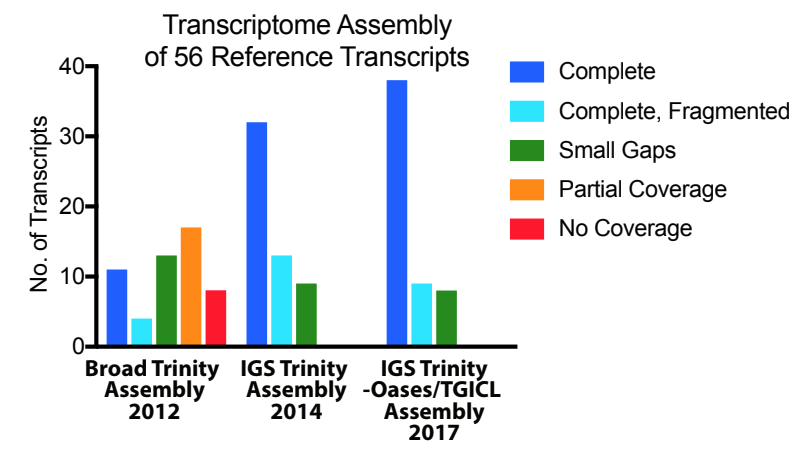
The improvements of the 2014 CNS assembly compared with a pilot transcriptome are illustrated in the figure below. Despite the overall high quality and completeness of this Trinity assembly, some transcripts remain fragmented, i.e., represented by two or more contigs.
Recent progress. We have used the Oases assembler with longer k-mer sizes (57- 85), in parallel with the Trinity assembler, and found some improvements in the combined transcript set, with increased coverage of some fragmented transcripts. We have generated combined contigs, using TGICL. Prior to running TGICL, spurious transcripts, e.g. chimeras, were removed, and contigs with gaps in read support were split. Various resulting assemblies are available on the new Aplysia Gene Tools blast site:
These transcriptome assemblies have not yet gone through NCBI contamination screens and could contain vector or other contamination.
In 2019, we plan to sequence the Aplysia genome on the new PacBio Sequel 6.0 platform. We expect that the long reads will enable a substantially more complete genome assembly than the previous Broad assemblies. This new genome assembly will be used in combination with the combined Trinity/Oases assembly to generate a complete annotated set of Aplysia transcripts.
This Illumina sequencing was supported by grants from NSF and NIMH. The data analysis and this website are supported by an EAGER grant from NSF.
Links
Current state of transcriptome assembly
gate.io
Recommended approach to searching Aplysia transcriptome
Pilot IGS assemblies
Early BROAD assemblies
Progress in CNS transcriptome assembly
For any questions or feedback please email Aplysia.RNAseq@gmail.com